Write more small tools
Preprocessing LICOR Data with python (& uv)
You should write more small tools. At first this will be slower than doing the tasks manually but it’s wort the time so you can:
Sharpen your programming skills
Build the habit of breaking down problems into manageable chunks
Make subsequent automation efforts faster
Tasks that take a lot of time (either because they’re frequent or involved) or are critical to get right are great candidates for automation. Certainly it’s possible to go overboard and spend hours trying to automate a 30 minute task, but if you’re starting out in the lab I would encourage you to keep an eye out for automate-able tasks.
Here’s a recent example: Pre-processing LI-COR data.
To follow along you’ll need these files: - demoextraction_cli.py
The LI-COR machine provides us with a wealth of physiological measurements but the way it saves them is less than ideal.
When saved as a table, we actually get several sub-tables in a single excel sheet. 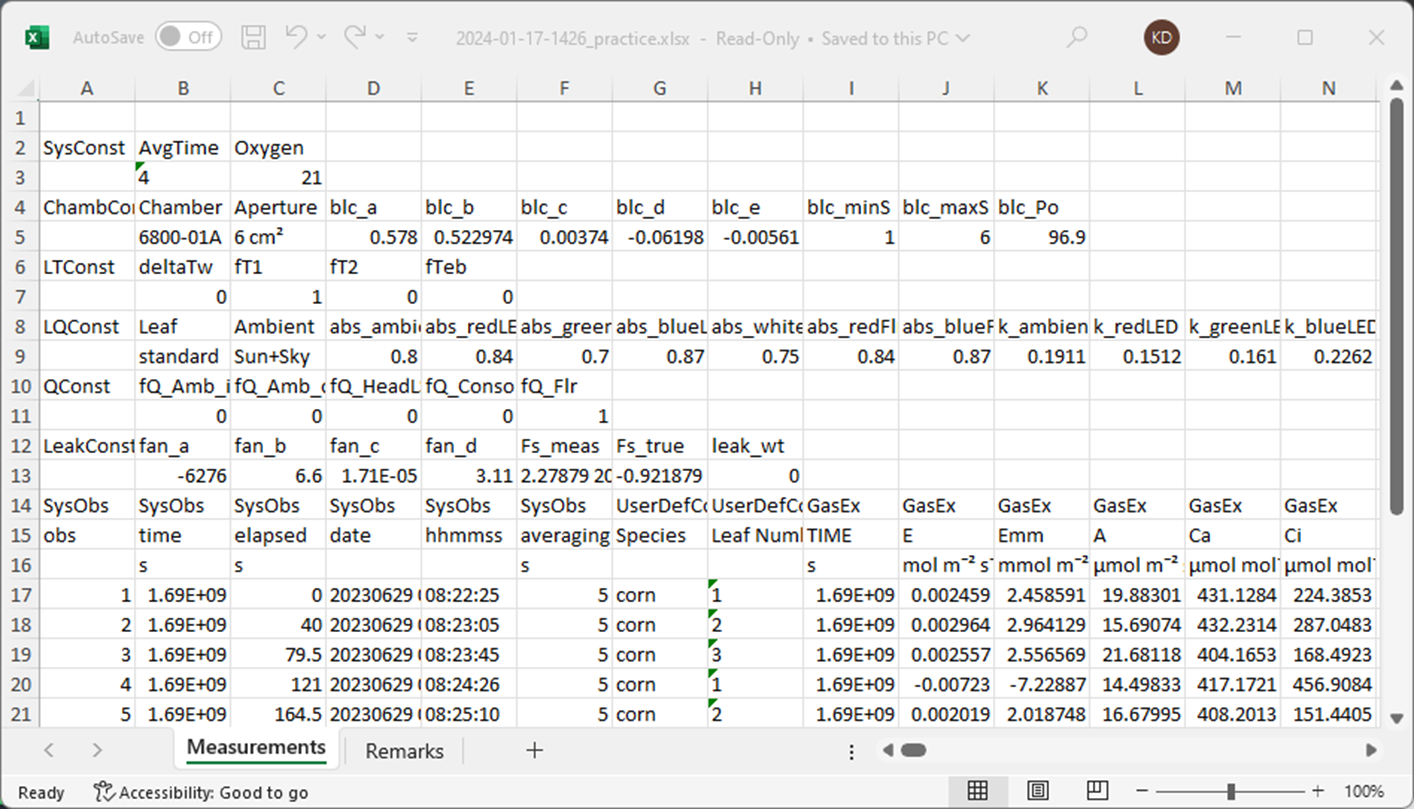
Here’s the other sheet in this file with metadata for the experiment. 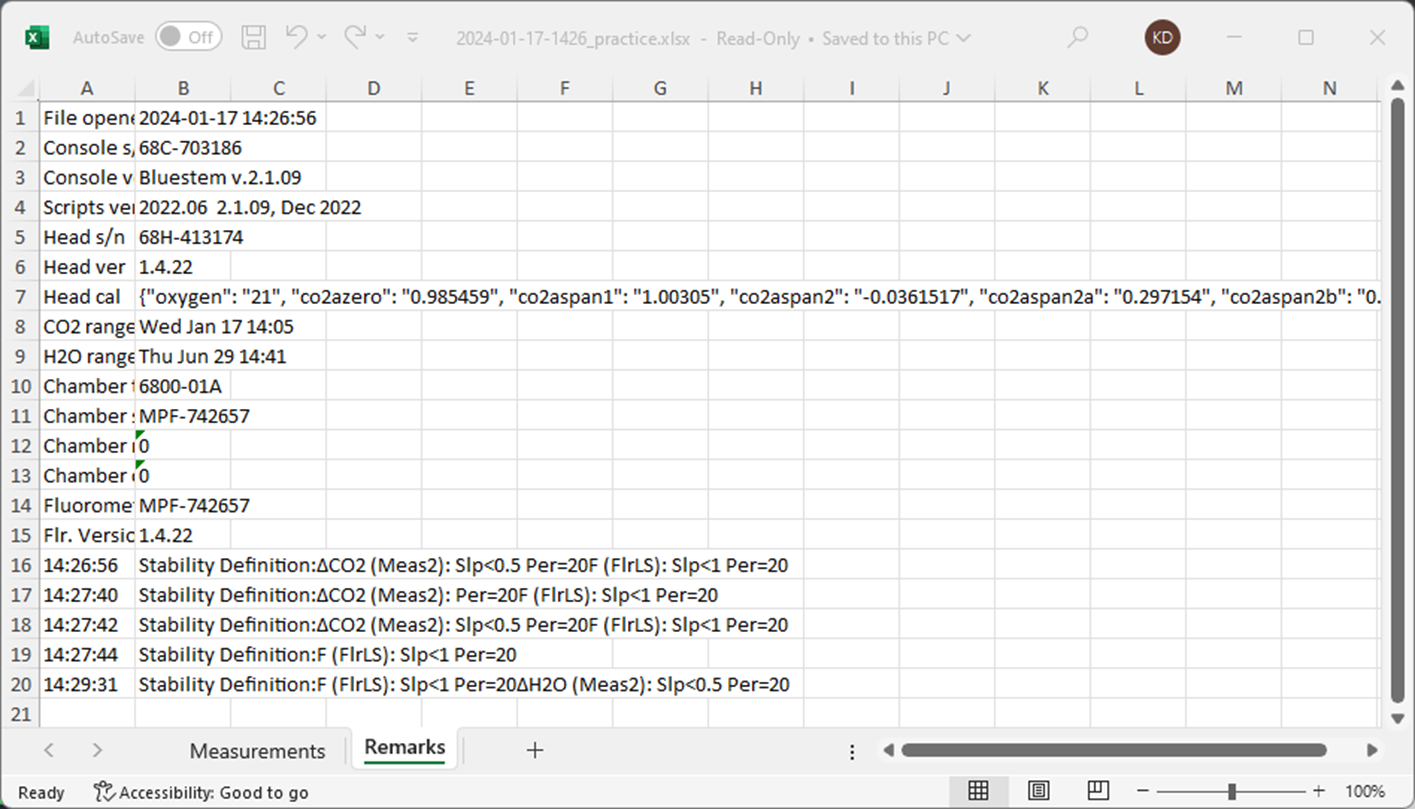
Importantly, we have one of these per use, so multiple sub-tables across files need to be aggregated prior to data analysis. Ideally we want something like this:
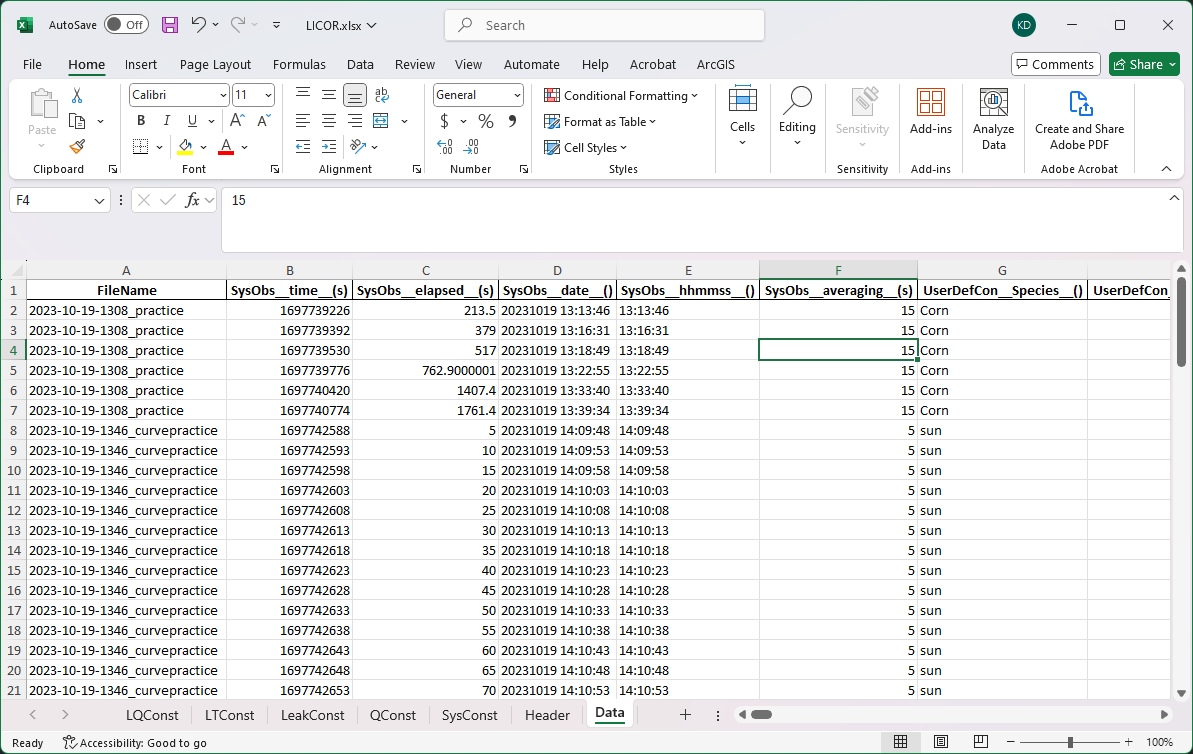
One file with data from multiple sessions and the data sorted from different types of metadata.
Okay, so how do we get from here to there? Well, we can work from these exported tables or from a text file that LI-COR uses. The text file’s are not as friendly for use but we can change that.
Here’s a glimpse at the file structure.
[Header]
File opened 2023-10-19 13:09:10
# ...
Head ver 1.4.22
Head cal {"oxygen": "21", "co2azero": "0.985459", # ...
"ssb_ref": "38488"}
CO2 rangematch Wed Oct 11 16:02
H2O rangematch Thu Jun 29 14:41
# ...
LeakConst:fan_d 3.11
LeakConst:Fs_meas 3.90048 209.364 364.041 589.972 873.062 1075.54 1262.06 1396.01
LeakConst:Fs_true -0.35308 225.423 382.151 585.793 808.815 1006.32 1201.68 1401.37
LeakConst:leak_wt 0
[Data]
SysObs SysObs # ...
obs time
s s
1 1697739012.6 0
2 1697739226.1 213.5
3 1697739391.6 379.0
# ...
7 1697740774 1761.4000000953674We have some tags ([Header], [Data]), entries that look like key/value pairs (File opened), dictionaries of key/value pairs (Head cal), arrays (LeakConst:Fs_meas), and a table of data.
The table is interesting because we have multiple headers – seemingly a grouping, variable name, and unit – before the data.
Conceptually, we could break this file into two chunks by the tags. We’ll sort all the metadata by the keys and store different kinds of data in their own table. The rows following the [Data] tag we’ll treat as a data table and combine all of the header information into a single header (SysObs time s becomes something like SysObs__time__(s)). Then we can save these data into an excel file and add new data to it.
I don’t want to dig into how the script achieves this. Rather I want to talk about how to get started building tools you want to use. We always start by thinking through assumptions and trade offs. In the above we assume that there is exactly one header and one data tag (this isn’t always the case!). We also trade some flexibility moving from a plain text format to an excel file (I think it’s worth it here but you might disagree!). There are also assumptions I’ve made that you’d have to look at the script to see. For instance, by default the script tries to process all in the working directory that do not have a file extension. To use it we could move the file to where the data is, start a virtual enviorment and run it like so:
(demo)$ python demoextraction_cli.py
Beginning:./2023-10-19-1308_practice
Beginning:./2023-10-19-1346_curvepractice
Beginning:./2024-01-17-1426_practice
Running DeduplicationWhat if we want more flexibility? Python has a number of libraries that let a script accept flags as arguments (I’m using the default one argparse). Now we don’t have to copy the script over and over, we can just change the input directory to the folders with the data.
(demo)$ python demoextraction_cli.py --inp_dir './' --out_dir './'What if we want to go even further? This script depends on a few common libraries, but maybe yours needs more. What happens if you want to share it?
For that case I recommend the python package manager uv. In addition to being lightning fast it allows for ad hoc specification of script dependencies.
$ uv run --with pandas --with openpyxl python demoextraction_cli.pyThis will create an ephemeral virtual environment for the script. It’s a little bit messy to type all this out every time which brings us to uv’s next killer feature.
We can use the following format to specify a scripts dependencies.
#!/usr/bin/env -S uv run --script
# /// script
# requires-python = ">=3.10"
# dependencies = [
# "pandas",
# "openpyxl",
# ]
# ///
import os, re, argparse
import pandas as pd
#...Now uv can get the needed information from the script rather than from the command line. We can run it and it will happily setup the environment and work through the files.
$ uv run demoextraction_cli.py
Reading inline script metadata from `demoextraction_cli.py`
Beginning:./2023-10-19-1308_practice
Beginning:./2023-10-19-1346_curvepractice
Beginning:./2024-01-17-1426_practice
Running Deduplication